Double Match Triangulator
Version 5.2.1, 12 Jul 2023
Double Match Triangulator (DMT) is an autosomal DNA analysis tool for Windows.
DMT automates the analysis of two or more segment match files with common ancestor information you supply.
DMT finds all triangulating segments that two people share with some third person.
The user adds known Most Recent Common Ancestrors (MRCAs) and DMT uses those
to determine ancestral paths for each segment and to put the people into ancestral clusters.
The results are given to you in Excel files with details for each match, and a summary of all people.
You also can have DMT produce a file containing ancestral paths than can be uploaded to DNA Painter.
Double Match Triangulator was imagined and developed by Louis Kessler (Behold Genealogy).
The concept of double matching to determine triangulation groups enabled
DMT to place 3rd in the Innovator Showdown at RootsTech 2017 in Salt Lake City.
The Purpose of DMT
Our goal as genealogists is to extend our family tree. We take DNA tests to try to find relatives who may share common ancestors with us. We know how we're related to some of the people our DNA matches to. But for most of our DNA matches, we don't know that. If we can figure out how we're related to the other people our DNA matches, we'll have new branches we can add to our tree.
Double Match Triangulator (DMT) is an autosomal DNA analysis tool to help you find how the people you DNA match with may be related.
DMT combines segment match data of two or more people to find all the double matches and triangulating segments they share. Using known common ancestors of known relatives, DMT determines ancestral paths to indicate the common line that the DNA passed through and it organizes these into triangulation groups for you. DNA relatives who match in any particular triangulation group may have a common ancestor along or beyond the ancestral path who passed this segment to all the people in the group. The assignment of ancestral paths to segments of your DNA is known as Chromosome Mapping or DNA Painting.
Over the past five years Tim Janzen and Jim Bartlett have developed various manual techniques for chromosome mapping. They are leaders in this field and have both been able to determine the triangulation groups that cover almost all their DNA. This work deals with a lot of data and they have done this manually in spreadsheets, taking a lot of their time and effort. Double Match Triangulator has been developed to automate this process, and to make this type of chromosome mapping possible for anyone.
Double Match Triangulator will produce the following output for you:
-
Every one of your matches is presented visually in an Excel spreadsheet. Every triangulation that is possible from the segment match data is shown. You will see how the segments overlap with each other. The ancestral line, if one can be determined, is shown for each segment. The segment matches are then organized into
Triangulation Groups. Segment match data is shown on the Chromosome Map pages.
-
Summary information for every person is provided in an Excel spreadsheet. It includes the ancestral lines that are determined for every match for each person. DMT assesses the ancestral lines to assign a primary ancestral line to each person. The people are organized for you by ancestral line, which in effect is doing a
DNA Clustering of your matches for you. Person summary information is shown on the People page or file.
3. Ancestral path data for input to the DNA Painter website is optionally provided in a .csv file. You can take this file and upload it to DNA Painter to add your ancestral paths from DMT to an existing or a new DNA Painter profile.
4. Log files are created after every run that give detailed information about the results, including number of people, number of segments, double matches and triangulations, parent and sibling information, persons A and B's matching segments, errors in the input files, as well as progress information about the run. Combined runs also include a segment size summary.
With DMT, you can skip the long and involved steps of looking for matching segments, identifying triangulations, identifying the ancestral path of each segment, and determining triangulation groups. Instead you'll be able to jump right ahead to using the results to help you identify how your DNA matches are related and extend your family tree.
Segment Match Data
DMT uses segment match data that is available from Family Tree DNA, 23andMe, MyHeritage DNA, or GEDmatch. These are the only companies that provide you with all your segment match data. AncestryDNA does not give you access to your segment match data. To use your AncestryDNA results you have to upload your DNA raw data to Family Tree DNA, MyHeritage DNA or GEDmatch and then download your segment match data from there. GEDmatch requires a Tier 1 subscription ($10 to $15 for a month) to access the utility to download segment match data from their site.
Segment match data differs between companies. You cannot compare segment match data from one company with segment match data from another company. The main reason is that matches need to compare to the same person in both files, but the same person often didn't test at different companies and when they did may have used different display names at the different companies. To a computer, John Smith is not the same as J. Smith or John D. Smith or JS. An additional reason is that each company uses their own proprietary algorithm to determine whether two people match over a segment and where the start and the end of a match is. Using two files from the same company eliminates differences due to matching algorithms.
Although DMT will accept just one segment match file, you cannot determine triangulating segments from just one file. So you really will want to use two or more segment match files from a single company to get full value from DMT's analysis.
Your own segment match file contains all the segments of all the people you match to. You'll want segment match files from some of the people you match to that you can compare with.
Here are some of the people you would like segment match files for:
-
Cousins whose relationships you know. If you know your common ancestors with your cousin, then most of your segments in common with that cousin should match through those common ancestors.
-
Aunts/Uncles, half siblings. They will match one of your parents and both your grandparents on that parent's side, helping you partition your father's matches from your mother's.
-
Parents. Matching with a parent is what I call parental filtering. If a parent doesn't match on the same segment that you match someone, then that match is either on your other parent, or it is a false match. DMT gives you additional information for parent matches. If you have both your parent match files, then any of your matches that neither parent matches will be shown by DMT as likely being a false by-chance match.
-
Siblings. They are tricky, because you can match them on both parents at the same time. But they are still very useful for double matching and, are quite usable for triangulations and Missing A-B and Missing B-C matching. In addition siblings are excellent for inferred matching, which is where your sibling matches someone but you don't.
-
Nephews/nieces. They only get one chromsome from your sibling, so they won't match on both your parents at the same time. They should match to many of your relatives on both your father and mother's sides.
-
Children/grandchildren. Although they can only match on segments that you match, they still are very useful for triangulations since the child effectively phases the parent to one set of alleles which then match a third person.
-
Someone who you know is related but don't know how. The cluster that these people are put into may be able to help you identify your common ancestor with this person.
-
Children of matches. They may allow you to triangulate with their parent.
-
Anyone else who matches you. You never know what might be revealed.
Important Concepts
Autosomal DNA
Each of us have 23 pairs of chromosomes, one of each pair from our father and one from our mother. The 23rd pair are two X chromosomes for a woman, and an X and a Y chromosome for a man. The Y chromosome is not included as part of autosomal DNA, so a man will only have a single X in the 23rd position for the purpose of autosomal DNA analysis.
Above is an example showing the 45 autosomal chromosomes for a man drawn with DNAPainter. The chromosome pairs are numbered 1 to 23, with number 23 known to be X. The top chromosome of each pair is from the person's father and the bottom of each pair is from the person's mother. The single X chromosme for a man is from his mother.
Recombination and Crossovers
Each parent passes you 23 chromosomes. Like you, your parent has 2 of each of their chromosomes, and in the process of giving you yours, they recombine each of their pairs into one for you. It is done in a simple manner, by taking the first bit from one parent, the next from the other, back to the first, etc. Each chromosome normally "crosses over" only a few times. So parts of the chromosome are from one grandparent, and parts are from the other. It is possible for a chromosome not to crossover, and in that case, the whole chromosome will be from one grandparent. The above diagram shows light and dark green segments of DNA that are likely from the person's patenal grandmother and blue segments that are likely from the person's paternal grandfather.
Ancestral Paths
Each little bit of DNA comes down some ancestral path. You get it from either your father or mother. Your father or mother gets it from their father or mother, etc. DMT will use "F" for father and "M" for mother to denote an ancestral path. So FMF comes from your father's mother's father, i.e. your great-grandfather. This Ancestral Path helps you to figure out how a relative may be related to you. If a person matches you on, say, an FMF segment, then they may be related to you on your father's mother's father's side. Using that information, you can look for records that may show your connection through that ancestral path.
Most Recent Common Ancestor (MRCA)
When two people are related, they share common ancestors. The Most Recent Common Ancestors of Person A and Person C are those who are the closest two ancestors who may have passed a segment to both A and C. For example, if A and C are second cousins, then they share a pair of great-grandparents. You will need to assign the MRCA to Person C and save it in DMT's People File so that DMT can use it. The MRCA is similar in contruction to an ancestral path, but it usually ends in an "R" to represent a paiR of ancestoRs, e.g. your great-grandfather and great-grandmother. So an MRCA for your 2nd cousin might be: FMFR, indicating that your father's mother's father's parents are also your cousin's great-grandparents. DMT uses the MRCA information you supply to create ancestral paths for each segment. So DMT can only go back as far back as the MRCAs you supply to it.
Segment Match Data
The data from each company that DMT uses is different from each other, but generally includes the following information:
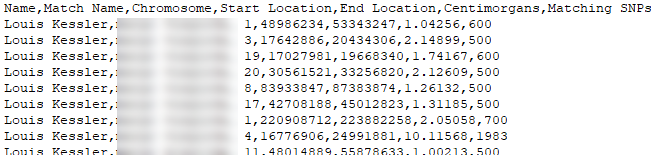
1. The name of the DNA tester, we'll call Person A
2. The name of another DNA tester who Person A matches to, we'll call Person C.
3. The chromsome they match on.
4. The starting position on the chromosome of the match, in base pairs (bp).
5. The ending position on the chromosome of the match, in base pairs (bp).
6. The genetic distance of the match in centimorgans (cM).
7. The number of tested Single Nucleotide Polymorphisms (SNPs) on that segment.
Each company tests the DNA at their own set of SNP locations. They each use their own proprietary method for determining what they will call a segment match between two people.
Single Matching
The information in one segment match file gives all of a person's single matches. There are two values at each SNP, one for the father and one for the mother. The DNA test results cannot differentiate which value at each SNP is the father's and which is the mothers. As a result, the testing companies compare both of Person A's values at a SNP to both of Person C's values at each SNP. If either of A's values match either of C's values, then that SNP is called a match. The more SNPs that match on this basis that you have in a row, the more likely it is that one chromosome on A is matching one chromosome on C. Once you reach a certain number of SNPs, often thought to be about 3000, equivalent to about 15 cM, then the likelihood of a match by chance or false match, where either chromosome of A is matching either chromosome of C, becomes very small. If this is not a match by chance, then you can be fairly certain that one of Person A's parents matches one of Person C's parents over the majority of the segment. Assigning matches to one parent is called phasing. Two phased matches are often Identical by Descent (IBD) where a common ancestor has passed the segment down to both Persons A and C. If you know how Persons A and C are related, then you may know who their common ancestors are, and you can map this segment to the ancestral path leading to these common ancestors. This ancestral path can be given a color and "painted" onto the segment with a program like DNA Painter. The important thing to remember about overlapping single matches is that some may be on dad's side and some on mom's. Just because they overlap, you cannot assume they are on the same side without additional information.
Double Matching
When you have segment match data for a second DNA tester, we'll call Person B, then you can compare Person A's matches with every Person C to Person B's matches with every Person C. If Person A and Person B both match the same Person C on the same chromosome and the start and end locations match or overlap each other, then the overlapping part will be a double match. A double match by itself does not mean much by itself. It needs a bit more.
Triangulating Segment
When you have three people, Person A, B and C, and Person A matches Person C, Person B matches Person C, and Person A matches Person B, all on the same chromosome and their start and end locations all overlap, then the common overlapping segment is called a triangulating segment. Because you now have 3 people in pairs all matching each other, it takes fewer SNPs to make the a match by chance unlikely. Only about 1400 SNPs or 7 cM are now needed to be fairly certain that one of each of Person A, B and C's parents all match each other over the overlapping part of the match. The other thing about three people who triangulate on a segment is that the three people generally will match on the same chromosome, either your father's or your mother's. (There are a few exceptions to this, e.g. two or more of the people being siblings, people related more than one way, or triangulations on opposite parents). A trinagulating segment will often (but not always) have been passed down to to Persons A, B and C from a common ancestor of the three.
Triangulation Groups
When Person A, Person B and Person C triangulate on a segment, then they form a triangulation group. If other people also match Person A, Person B and Person C on the same segment, then they are also part of the triangulation group. In actuality, every person in the triangulation group must match each other. But in practice some may not if their total shared matches did not reach the company's minimum or if one person's match file was downloaded before the other's test results came in. A matching segment between two or more people has three possibilities:
Double Match Triangulation
Double Match Triangulator works by using two match files of Person A and Person B. It finds all double matches that A has with C and B has with C on the same segment. Person A's match file also has matches with Person B (as does Person B's with Person A) and now it is a simple matter to see if Person A and Person B match on the same segment. If they do, we have a segment triangulation. Double Match Triangulator thus can easily find all the segments and people that Person A and Person B triangulate with. The discovery of this led to the development of DMT.
Identical By Descent (IBD)
If a segment is passed down to two or more people, that segment match is called Identical By Descent (IBD). This also means that when two or more people share an IBD segment, they share a common ancestor who passed that segment down to them. If the segment data is corrent, then every person that shares the same IBD segment should triangulate on that segment. The reverse is not always true, as triangulation does not always mean IBD (see A Few Caveats, below).
In Common With (ICW)
When two related people, Person A and Person B, both share DNA with a Person C, they are said to be sharing DNA In Common With Person C. This does not necessarily mean that they share overlapping segments or triangulate anywhere. But it does mean that Person A and Person C are likely related, and that Person B and Person C are likely related. All DNA companies provide a list of In Common With people. Ancestry DNA does not provide segment matching data, but does provide ICW people that can be used with various clustering techniques (see Clusters, below).
Missing AB, Missing BC
In Common With information is very important in Double Match Triangulator. When you have a double match where Person A matches C on a segment and Person B matches C on the same segment, it is very important that they all share data with each other somewhere, i.e. that they are ICW each other. So on that double match, if Person A also matches Person B on that same segment, we have a triangulating segment. But if Person A and B don't match each other on that segment, but do match each other elsewhere, we then have what we call a Missing AB match. Similarly, if Person A matches C on a segment, Person A matches B on that segment, Person B does not match C on that segment, and A, B and C are ICW, then we have what we call a Missing BC match. DMT shows all Missing AB and Missing BC matches in its analysis.
Inferred Matches
An inferred match is where Person B matches Person C on a segment but Person A does not match Person C on the same segment. Persons A, B and C must be ICW each other. If you know how Person A and B are related, and how Person A and C are related, then the segment where Person B matches Person C might tell you what A's segment's ancestral path cannot be. DMT finds all possible inferred matches and displays and uses them in its analysis.
Clusters
You can group people into clusters from lists of ICW people using techniques such as the Leeds Method and clustering programs based on it. Clustering people is very useful. When the relationships of a few people in a cluster are known, they can be indicative of what the ancestral path might be for all people in the cluster. DMT puts people into clusters as well, but rather than using ICW information at a person level it instead uses the ancestral paths of the segment matches of the person. Different clustering tools provide different resulting clusters, and DMT provides its own cluster results for you to make use of.
A Few Caveats
Three or more overlapping IBD segments should triangulate with each other. However, sometimes they may not because:
Triangulation does not necessarily mean IBD, because:
-
One of the segments may match by chance to the other two, expecially if small (e.g. under 7 cM)
-
Three triangulating segments may match each other each on opposite parent chromosomes, e.g. (1) paternal A matches maternal B, (2) paternal B matches maternal C, and (3) paternal C matches maternal A. In this case, there are three different common ancestors each passed down a segment to two of A, B and C.
-
The company's algorithm, imputation, read errors or mutations, may result in a triangulation where there really is not one.
In other words, if you have a triangulation, don't assume it is IBD.
But, because an IBD segment shared by 3 or more people must triangulate, a segment that triangulates passes the first test that it might be IBD.
Nothing Is Guaranteed
DMT must listen to what the data is telling it. It has to assume that the segment matches it is given are correct. If the data says that 3 segment matches don't triangulate, then DMT has to accept that. Due to the caveats specified above, some of the data DMT makes use of could be incorrect. Some of the incorrect data may affect the results.
You will be entering into DMT, the common ancestor information for your DNA matches whose relationships you know. Most of the segments you match with a person will likely be along the MRCA you specified. Some segments, however, may come from other lines if that relative is related more distantly on some other line. Or maybe you're from an endogamous population and some segment matches come from who-knows-where. These segments that are not from MRCAs you specify will therefore be incorrectly assigned by DMT. There will be some matching segments through closer relationships and others through more distant relationships. DMT uses "majority rules" as its resolution technique, and will accept the ancestral path that the most matches agree on. The correct relationships in theory should outnumber the incorrect relationships.
But not always. Be very aware that some results from DMT may not be correct. As you add more match data and more MRCAs that you know, DMT's segment assignments will improve.
DMT automates the analysis of more data than you'll ever be able to analyze by hand yourself. Just remember, as Blaine Bettinger said at MyHeritageLive 2019: "Automation does not give you the answer. It gives you a suggestion." Use DMT's suggestions wisely.